| Index to this page |
Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single recombinant molecule.
5' GGATCC 3'
3' CCTAGG 5'
To be useful, the recombinant molecule must be replicated many times to provide material for analysis, sequencing, etc. Producing many identical copies of the same recombinant molecule is called cloning. Cloning can be done in vitro, by a process called the polymerase chain reaction (PCR). Here, however, we shall examine how cloning is done in vivo.
Cloning in vivo can be done inIn every case, the recombinant DNA must be taken up by the cell in a form in which it can be replicated and expressed. This is achieved by incorporating the DNA in a vector. A number of viruses (both bacterial and of mammalian cells) can serve as vectors. But here let us examine an example of cloning using E. coli as the host and a plasmid as the vector.
| Electron micrograph showing plasmids (21K) |
Plasmids are replicated by the same machinery that replicates the bacterial chromosome. Some plasmids are copied at about the same rate as the chromosome, so a single cell is apt to have only a single copy of the plasmid. Other plasmids are copied at a high rate and a single cell may have 50 or more of them.
Genes on plasmids with high numbers of copies are usually expressed at high levels. In nature, these genes often encode proteins (e.g., enzymes) that protect the bacterium from one or more antibiotics.
Plasmids enter the bacterial cell with relative ease. This occurs in nature and may account for the rapid spread of antibiotic resistance in hospitals and elsewhere. Plasmids can be deliberately introduced into bacteria in the laboratory transforming the cell with the incoming genes.
5' GGATCC 3'that, as we saw above, is cut by the restriction enzyme BamHI
3' CCTAGG 5'
5' AAGCTT 3'that is cut by the restriction enzyme HindIII
3' TTCGAA 5'
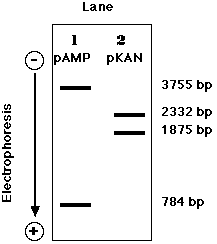 Treatment of pKAN with a mixture of BamHI and
HindIII produces:
Treatment of pKAN with a mixture of BamHI and
HindIII produces:
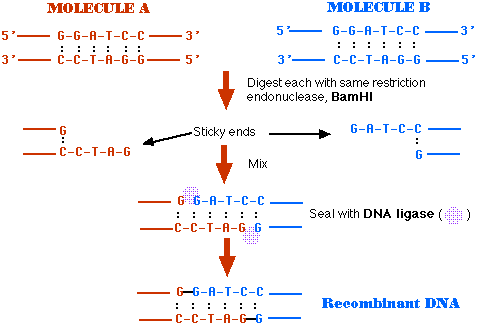
If you remove the two restriction enzymes and provide the conditions for DNA ligase to do its work, the pieces of these plasmids can rejoin (thanks to the complementarity of their sticky ends).
Mixing the pKAN and pAMP fragments provides several (at least 10) possibilities of rejoined molecules. Some of these will not produce functional plasmids (molecules with two or with no replication origin cannot function).
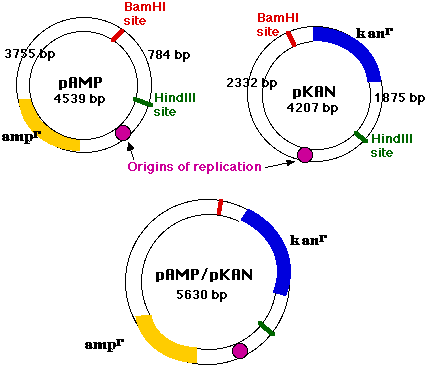
One interesting possibility is the joining of
Sealed with DNA ligase, these molecules are functioning plasmids that are capable of conferring resistance to both ampicillin and kanamycin. They are molecules of recombinant DNA.
Because the replication origin, which enables the molecule to function as a plasmid, was contributed by pAMP, pAMP is called the vector.
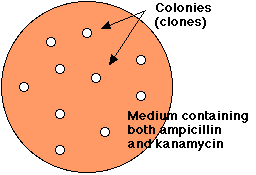 Treatment
of E. coli with the mixture of religated molecules will produce some colonies
that are able to grow in the presence of both ampicillin and kanamycin.
Treatment
of E. coli with the mixture of religated molecules will produce some colonies
that are able to grow in the presence of both ampicillin and kanamycin.
However, E. coli can be simultaneously transformed by more than one plasmid, so we must demonstrate that the transformed cells have acquired the recombinant plasmid.
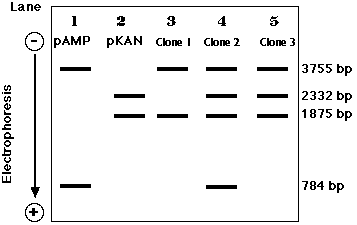
Electrophoresis of the DNA from doubly-resistant colonies (clones) tells the story.

5' GAATTC 3'This is cut by the restriction enzyme EcoRI, producing sticky ends.
3' CTTAAG 5'
If we treat any other sample of DNA, e.g., from human cells, with EcoRI, fragments with the same sticky ends will be formed. Mixed with EcoRI-treated plasmid and DNA ligase, a small number of the human molecules will become incorporated into the plasmid which can then be used to transform E. coli.
But how to detect those clones of E. coli that have been transformed by a plasmid carrying a piece of human DNA?
The key is that the EcoRI site is within the kanr gene, so when a piece of human DNA is inserted there, the gene's function is destroyed.
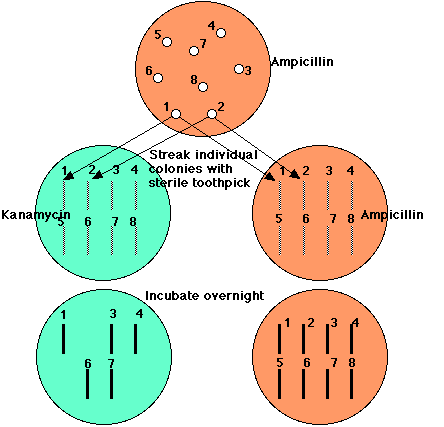
All E. coli cells transformed by the vector, whether it carries human DNA or not, can grow in the presence of ampicillin. But E. coli cells transformed by a plasmid carrying human DNA will be unable to grow in the presence of kanamycin.
So,All those clones that continue to grow on ampicillin but fail to grow on kanamycin (here, clones 2, 5, and 8) have been transformed with a piece of human DNA.
Using procedures like this, many human genes have been cloned in E. coli or in yeast. This has made it possible - for the first time - to produce unlimited amounts of human proteins in vitro. Cultured cells (E. coli, yeast, mammalian cells) transformed with the human gene are being used to manufacture:
| Welcome&Next Search |